As a bioinformatician, I remember the exact research question that started my journey: “Can we predict the function of hypothetical proteins from the human genome using only computational methods?” The problem was, I had no clear road map. My first year and a half felt less like a scientific investigation and more like a technician’s checklist. My days were a cycle of running a sequence through BLAST, then trying a multiple sequence alignment with ClustalW, then searching a database for protein domains. I was just trying things, hoping for a lucky hit. There was no strategy, just a frustrating process of trial and error. I was drowning in data but starved for a clear direction.
This struggle is exactly why I built the Literature Survey AI. I needed a tool that could give me the big picture first, transforming the mountain of existing research into a clear map so I could navigate it with purpose. You can try it out for yourself here, I designed it using an intuitive vibe coding method so it would feel like a helpful guide. To prove it worked, I gave the app the very topic that had once left me so lost.
Step 1: A Clear Starting Point, Not a Confusing Mess
When I started my research, I spent months just trying to grasp the basic concepts. The app’s first output, an Introduction, laid out the entire foundation in a few clear paragraphs. It defined what a hypothetical protein was and explained that using computers was the essential first step to study them.
Reading this, I felt a sense of relief. This simple summary would have saved me so much time and self-doubt. It provided the solid ground I had to spend months searching for on my own.
Step 2: A Strategic Plan, Not Just a Box of Tools
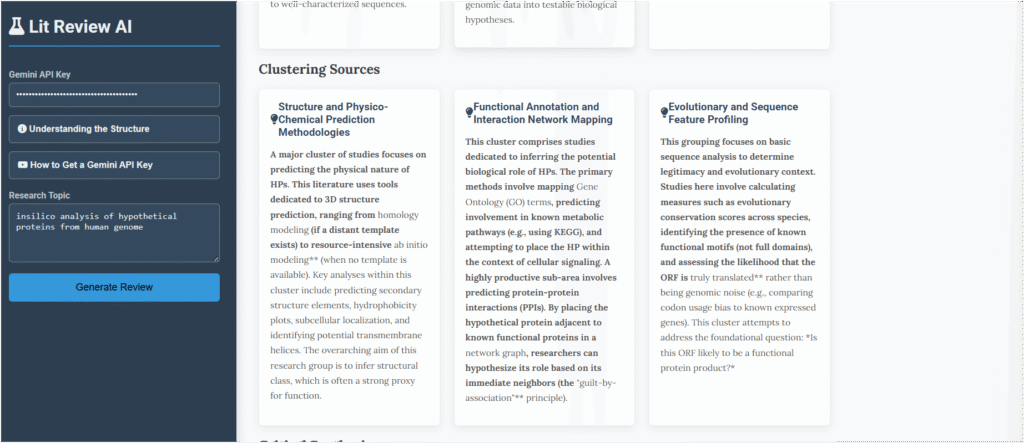
My initial approach was chaotic. I jumped between different analysis methods without a real plan. The app’s Clustering Sources section fixed this immediately. It took all the different techniques and sorted them into logical groups.
- Looking at the Sequence: This grouped all the methods like BLAST and ClustalW that I was using. It put my “technician” tasks into a proper scientific context.
- Predicting the Shape: This group focused on guessing the protein’s 3D structure to figure out its job.
- Seeing the Social Network: This cluster was about placing the mystery protein into known cellular pathways to see what its function might be.
This simple organization was a game-changer. Instead of just trying tools randomly, I would have had a clear, structured plan from day one. It showed me why each approach was important and how they fit together.
Step 3: Finding the Real Scientific Questions
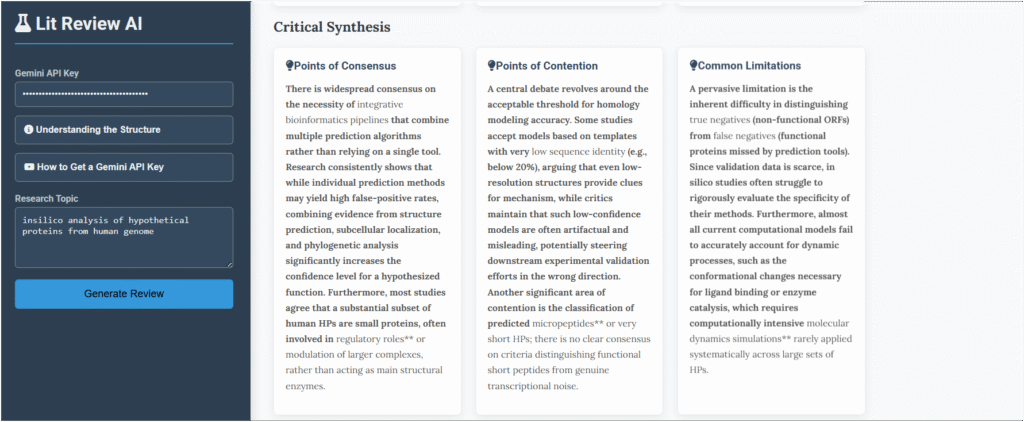
The Critical Synthesis section felt like talking to an experienced mentor. It didn’t just list facts; it explained what they meant.
- What everyone agrees on: It confirmed that using computers to make educated guesses about these proteins is the standard way to start.
- What people are still debating: It highlighted the big question—which computer method is the most reliable? This showed me where the real scientific conversation was happening.
- Common problems: It reminded me of the most important truth—all of this computer work is just a prediction. Nothing is real until it’s proven in a lab.
The Critical synthesis for the query ” Insilico analysis of hypothetical proteins from human genome ” is shown below:
Step 4: A Shortcut to New Ideas
The final section, Research Gaps, was the most powerful. It pointed to specific, advanced ideas that took me years to even learn about. It suggested looking into how these proteins might be involved in forming new structures in the cell (“phase separation”) or if they could have more than one job (“moonlighting proteins”).
Looking at this, I saw my 1.5-year struggle transformed into a clear path forward. I built this tool to be the guide I never had. My hope is that it can help other researchers skip the confusion and get straight to the part that matters most: discovery
Now, there’s one more thing I learned while testing my app that’s important to know. Because it’s powered by a generative AI, it’s not like a calculator that gives the same answer every time. If I run my topic—”in-silico analysis of hypothetical proteins”—multiple times, the results won’t be identical. I’ve found that the core structure and the main ideas remain consistent, but about 20% of the output might differ with each run. One time, it might highlight a slightly different “Research Gap,” or phrase a “Point of Contention” in a new way that sparks a fresh idea. This is why I see it not just as a summarizer, but as a brainstorming partner. Each run has the potential to offer a new perspective, helping you look at your research problem from a slightly different angle.
My Final Piece of Advice: Use It as a Map, Not a Destination
Now, a final and very important thought. As powerful as this tool is, we must remember what it is: a guide, not a final authority. The AI is incredibly good at seeing patterns and summarizing information, but it’s not a substitute for our own critical thinking.
Think of the output as a map showing you several possible trails up the mountain. The map can show you where the trails are, what the terrain might look like, and where others have gone before. But it’s still your job as the researcher to walk the path yourself. You need to read the primary research papers, verify the claims, and use your own expertise to decide which trail is the right one for your journey. The app gives you the possibilities; you provide the validation.